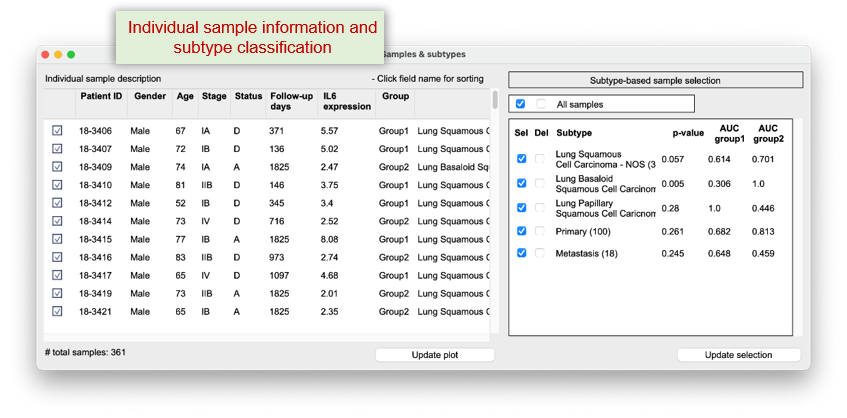
Powered by LLMs and Q-omics meta DBs, the OmixMind module enables users to perform flexible and intuitive data mining through natural language queries - offering greater versatility than menu-driven systems and requiring no bioinformatics or computational expertise.
The system automatically interprets user queries, generates appropriate analysis workflows and Python code, and initiates data mining.
To address the challenge of interpreting complex analysis results, an AI-powered interface further translates outputs into clear, comprehensive summaries and suggests new hypotheses - eliminating the need for data science knowledge.

* The OmixMind module is updated frequently, and an advanced version with user data integration will be available soon.
Demonstration
Type any questions
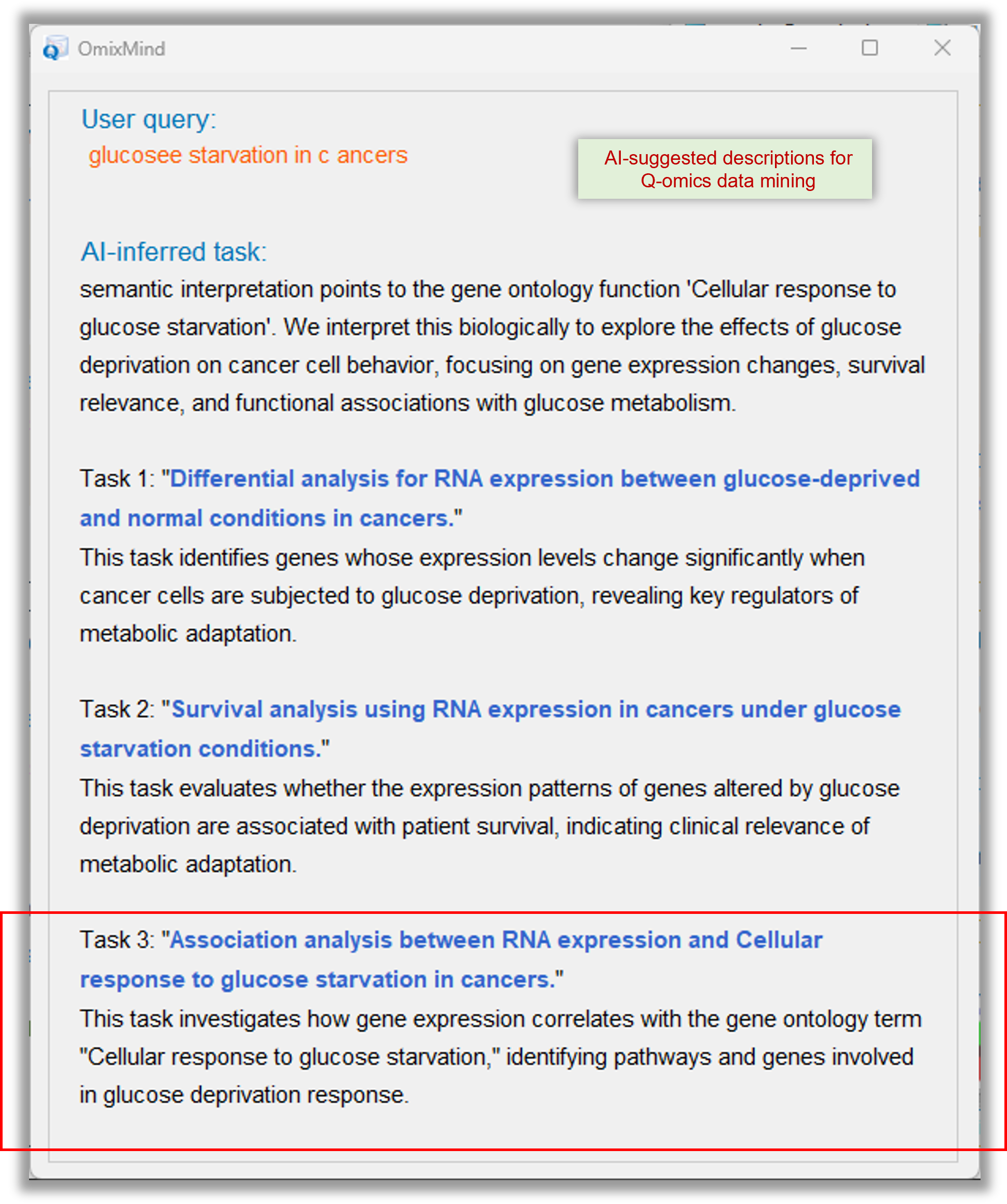
Example user query "glucose starvation in cancers" generates following workflow for Q-omics data analysis. Users can customize the workflow.
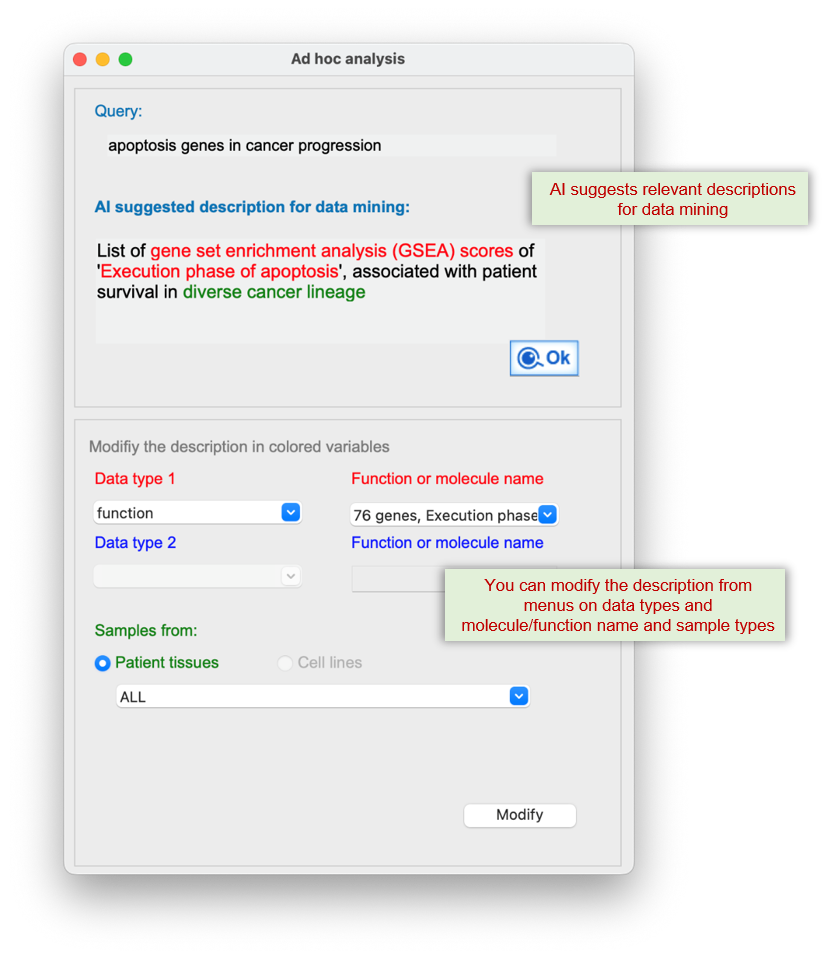
Significant data relevant to the query function and analysis type (i.e., patient survival), are retrieved from SmartDBs of Q-omics server.
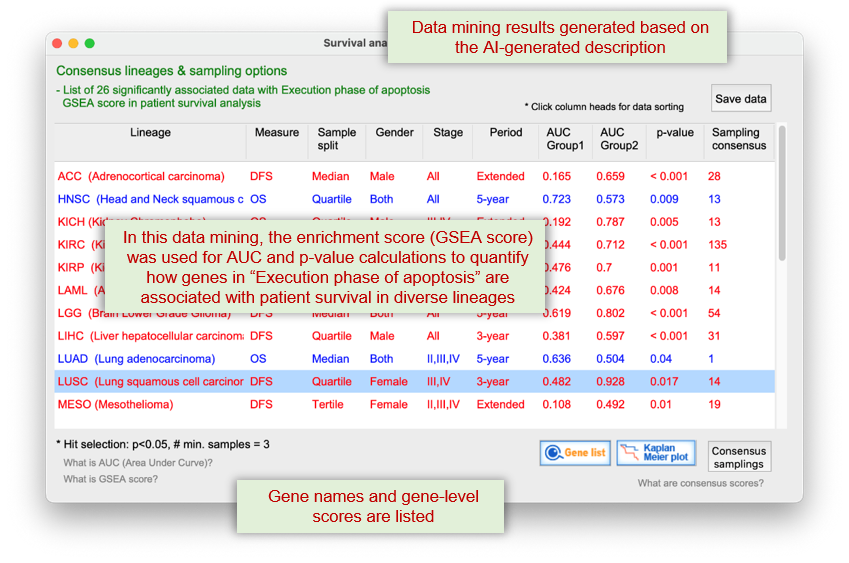
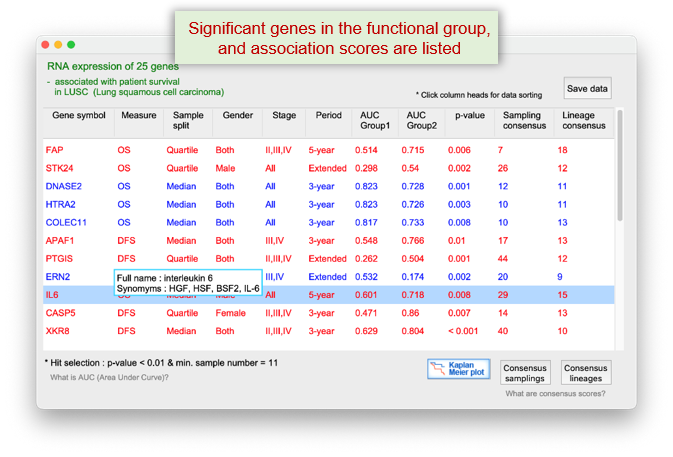
The "Gene list" button" in the panel above, displays the genes that significantly contribute to the enrichment of the query function in the survival of LUAD patients. The "NetCrafter" button below, provides the network-based functional analysis for the selected genes
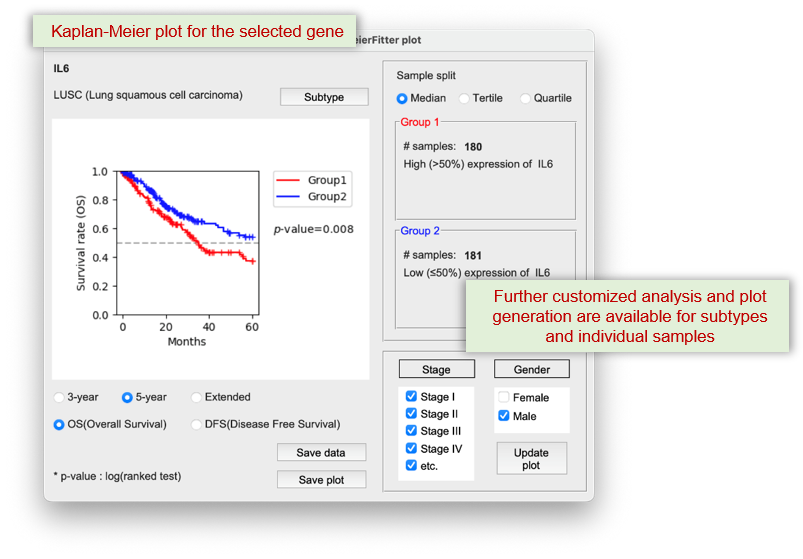
The 'Kaplan-Meier plot' button displays a graph of selected function or gene versus patient survival.
The NetCrafter-based network tool enables graphical analysis of the functional aspects of selected genes.