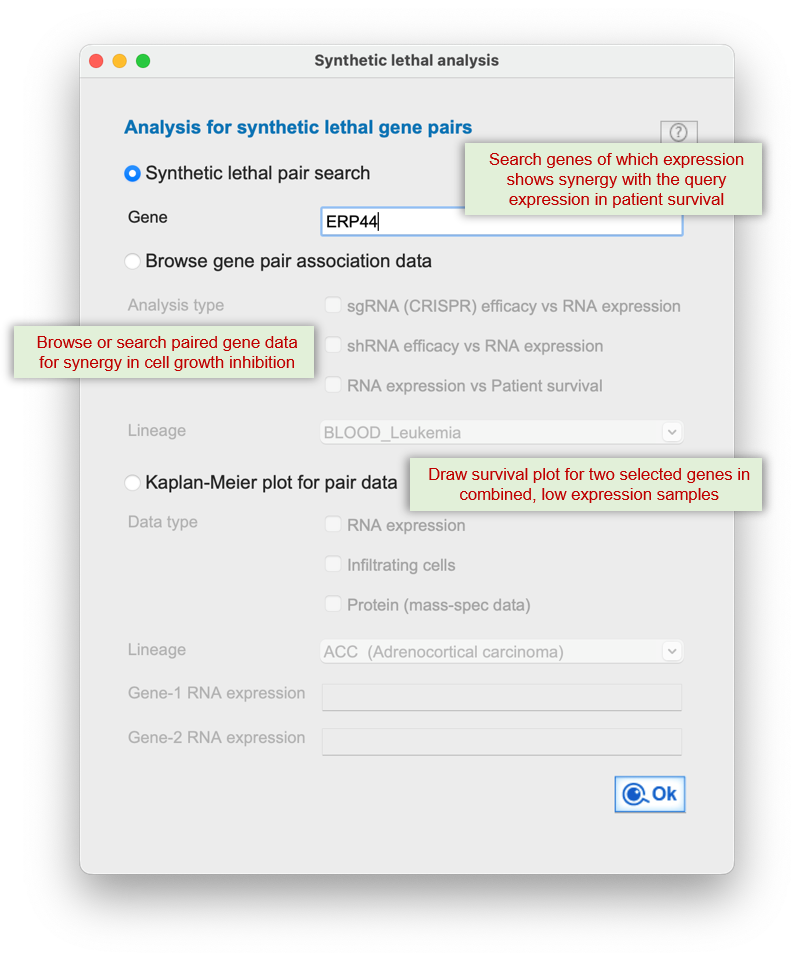
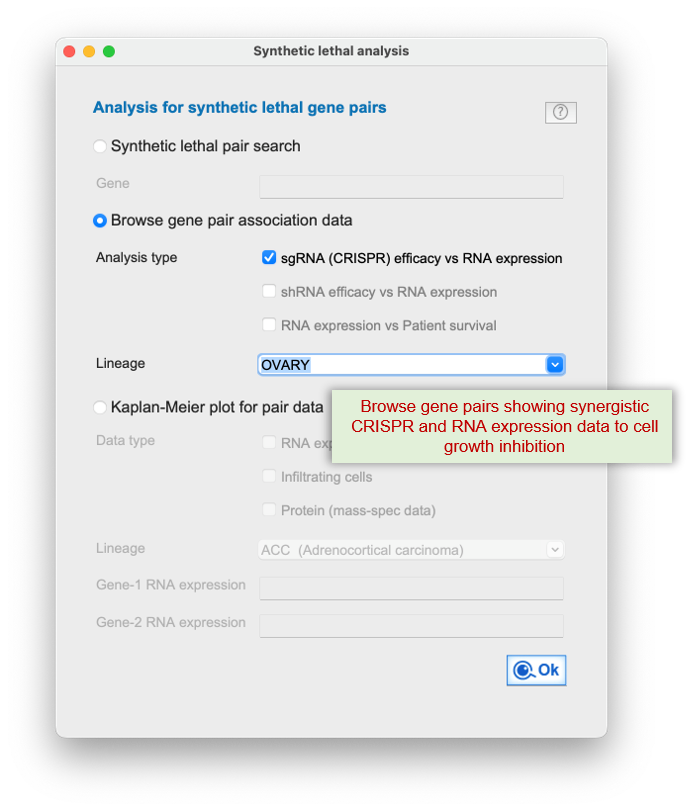
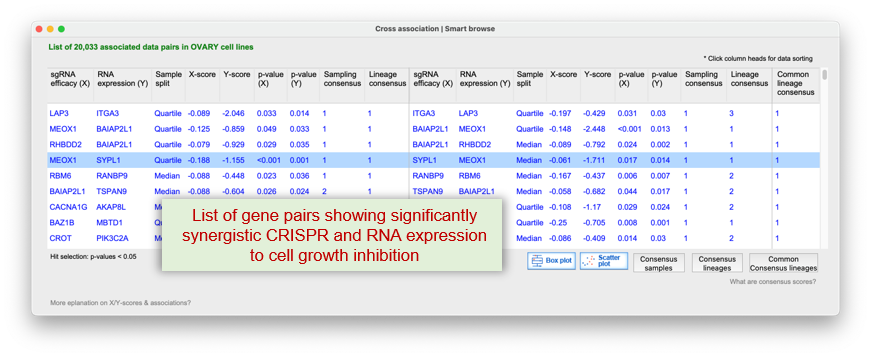
Q-omics integrates CRISPR knockout screens, RNA expression data, and patient survival outcomes to identify gene pairs whose
combined perturbation leads to enhanced cancer cell inhibition or improved clinical prognosis.
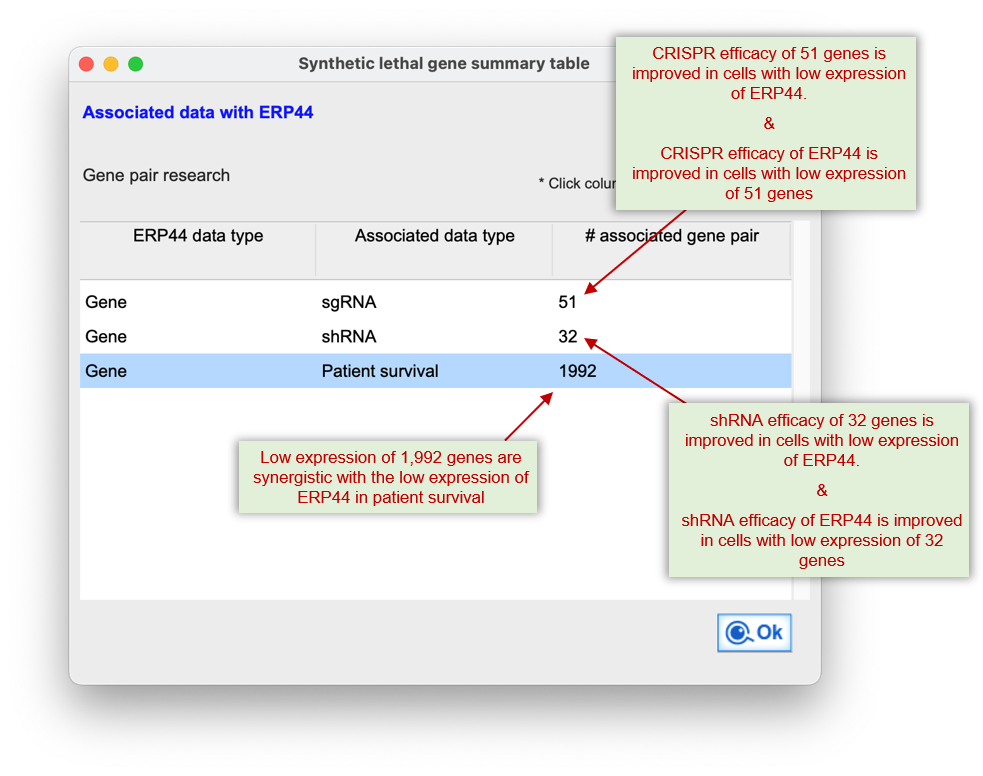
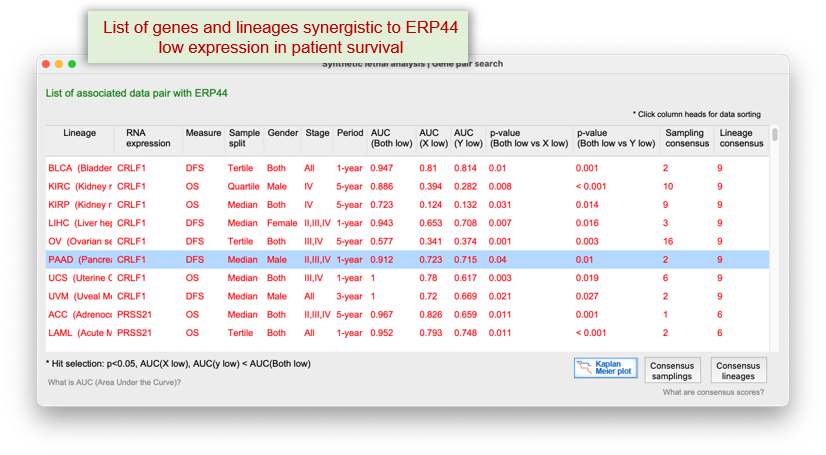
It systematically detects synergistic interactions where simultaneous disruption?such as one gene's low expression paired with CRISPR inhibition of another?induces stronger effects than single-gene targeting alone.
The platform also evaluates cases where co-downregulation of both genes correlates with better patient survival compared to single-gene downregulation.
This module emphasizes data consensus across cancer types, enabling the identification of robust and clinically actionable synthetic lethal targets for combination therapy.
It systematically detects synergistic interactions where simultaneous disruption?such as one gene's low expression paired with CRISPR inhibition of another?induces stronger effects than single-gene targeting alone.
The platform also evaluates cases where co-downregulation of both genes correlates with better patient survival compared to single-gene downregulation.
This module emphasizes data consensus across cancer types, enabling the identification of robust and clinically actionable synthetic lethal targets for combination therapy.
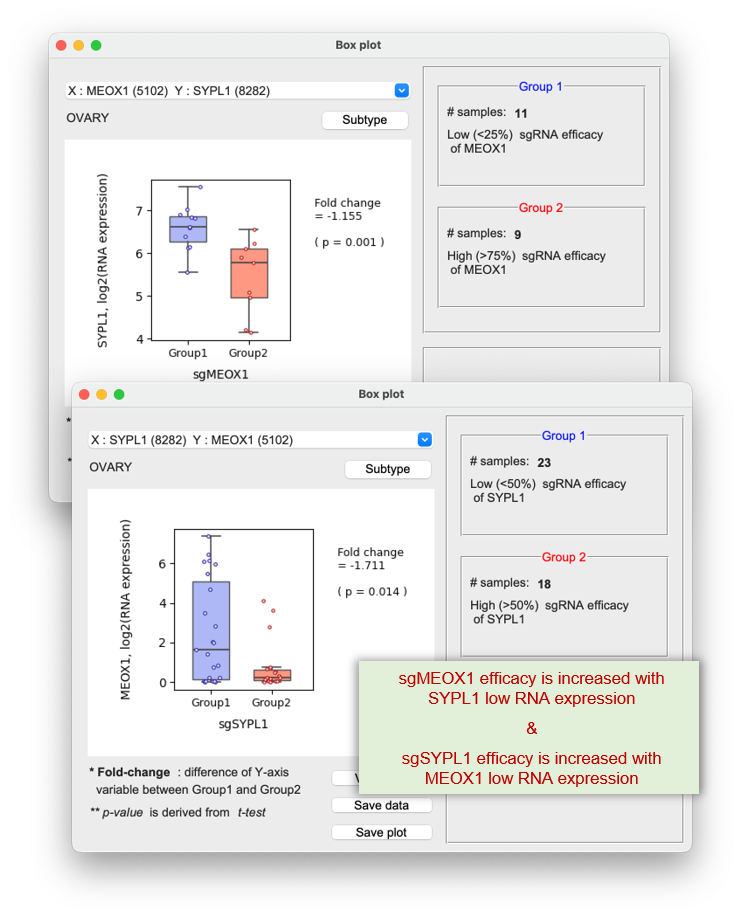
1) CRISPR (shRNA) efficacy of gene A are associated with mRNA expression of gene B, vice versa

Left panel) CRISPR efficacy of RNF113A is negatively associated with RNA expression of NKAP
Right panel) CRISPR efficacy of NKAP is negatively associated with RNA expression of RNF113A
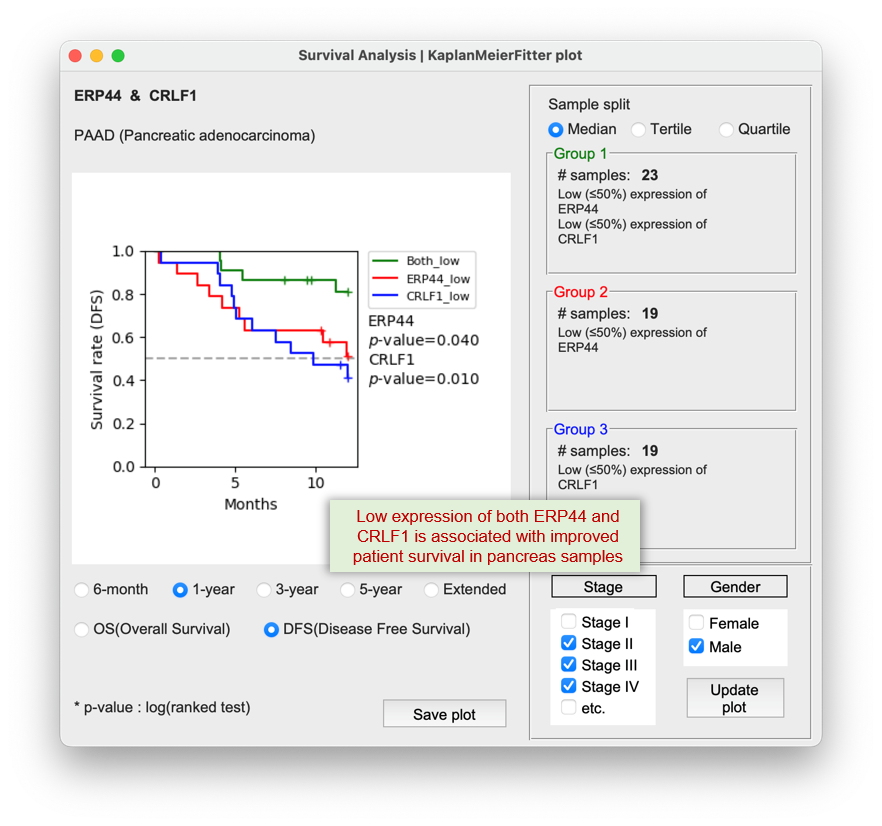
2) Low expression of both gene A & B are favorable to patient survival

Data mining workflows are available for evaluating the synthetic lethality of gene pairs