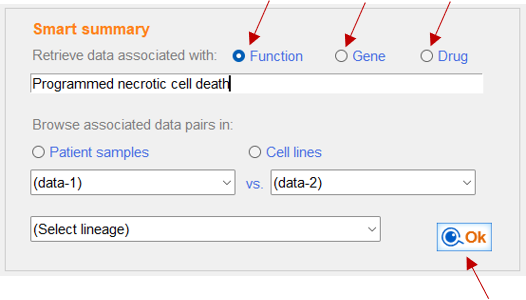
Smart DBs provides a list of all data and sample types associated with your query
|
Select query type: functional category, gene or drug name
|
Functional categories are >7,000 Biological process terms in Gene Ontology
All the associated data with the the selected functional category will be retrieved
GSEA score derived from RNA or protein data within the selected functional category is used for association analysis with other omics data
Gene selection will retrieve all associated data with your query across various data types: mutation, RNA, Protein (RPPA, mass-spec), CRISPR and shRNA
Drug selection will retrieve all associated omics data with your query drug
* 561 drugs are available
|
|
Programmed necrotic cell death is selected as functional query
|
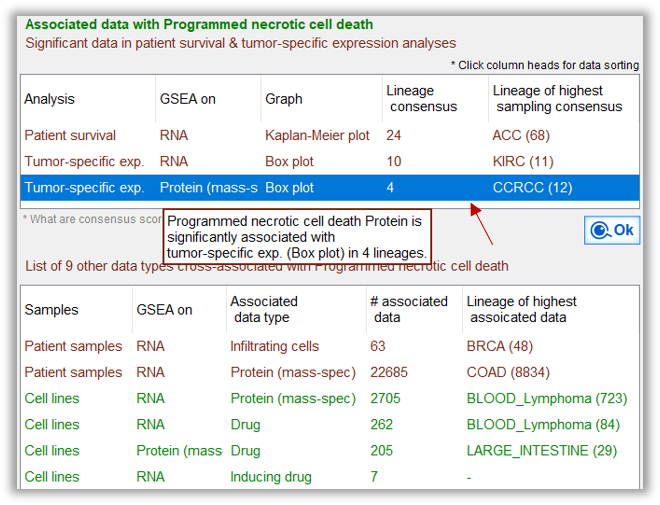
GSEA scores of RNAs and proteins within the functional category, are analyzed against patient outcome and other omics data Proteins in the query function is enriched in tumors of 4 cancer lineages |
|
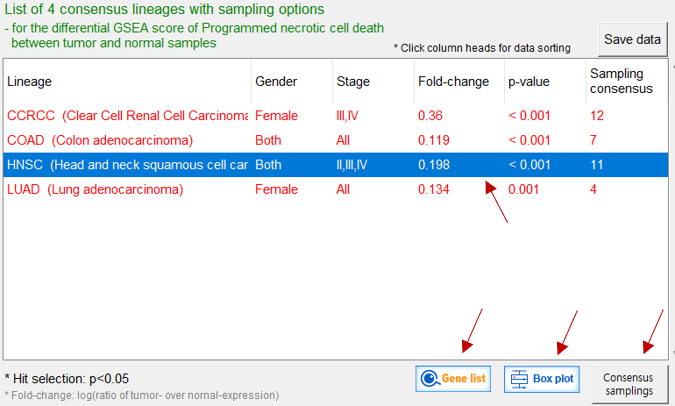
4 cancer lineages exhibiting significant enrichment (GSEA score) of proteins within the query function
|
P-value shows the significance of GSEA score fold-change between normal and tumor samples Gene list shows significant genes within the function Box plot display the selected data Consensus samplings show the 11 significant sample groups in HNSC |
|
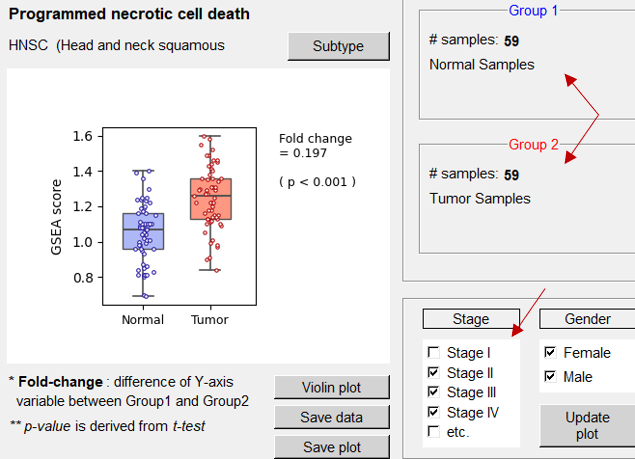
Box plot of GSEA score of query function in normal vs. tumor samples Proteins in the query function is significantly enriched in tumor compared to normal samples in HNSC patients
|
59 paired normal & tumor samples are compared in the plot Most significant result is observed in Stage II, III and IV samples in HNSC patients |